Overview of Datasets
Here, we summarize the T cell genomics datasets included in this study. These datasets cover genome-wide CRISPR screens, bulk and single-cell transcriptomic profiling of T and CAR-T cells, T cell-derived cancer data, and lymphocyte profiling data from autoimmune diseases.
T Cells CRISPR Screen Datasets
| PubMed | Year | StudyName | Species | CellType | Approach | Method | #Genes | Readout |
|---|---|---|---|---|---|---|---|---|
| 35276062 | 2022 | Ye_CRISPRa_CD8T | mouse | CD8T | in vitro | GoF | 18932 | kill |
| 35296855 | 2022 | Legut_ORFs_CD4T | human | CD4T | in vitro | GoF | 11608 | proliferation |
| 35296855 | 2022 | Legut_ORFs_CD8T | human | CD8T | in vitro | GoF | 11608 | proliferation |
| 35113687 | 2022 | Schmidt_CRISPRa_CD8T_IFNG_Prim | human | CD8T | in vitro | GoF | 18930 | IFNG |
| 35113687 | 2022 | Schmidt_CRISPRa_CD4T_IL2_Prim | human | CD4T | in vitro | GoF | 18930 | IL2 |
| 35113687 | 2022 | Schmidt_CRISPRa_CD4T_IL2_Supp | human | CD4T | in vitro | GoF | 18930 | IL2 |
| 35113687 | 2022 | Schmidt_CRISPRa_CD4T_IFNG_Supp | human | CD4T | in vitro | GoF | 18930 | IFNG |
| 35113687 | 2022 | Schmidt_CRISPRa_CD4T_TNF_Supp | human | CD4T | in vitro | GoF | 18930 | TNF |
| 36702900 | 2023 | Dai_CRISPRi_CART_CD4T | human | CART | in vitro | LoF | 899 | longevity and cytotoxicity |
| 36702900 | 2023 | Dai_CRISPRi_CART_CD8T | human | CART | in vitro | LoF | 899 | longevity and cytotoxicity |
| 32640256 | 2020 | Loo_CRISPRi_Treg_Expand | mouse | Treg | in vitro | LoF | 19668 | Expansion |
| 32640256 | 2020 | Loo_CRISPRi_Treg_Foxp3 | mouse | Treg | in vitro | LoF | 19668 | Foxp3 |
| 36550322 | 2023 | Szeto_CRISPRi_CD4T | mouse | CD4T | in vitro | LoF | 19674 | differentiation |
| 35113687 | 2022 | Schmidt_CRISPRi_CD4T_IL2_Prim | human | CD4T | in vitro | LoF | 18939 | IL2 |
| 35113687 | 2022 | Schmidt_CRISPRi_CD8T_IFNG_Prim | human | CD8T | in vitro | LoF | 18939 | IFNG |
| 32499641 | 2020 | Cortez_CRISPRi_Treg | mouse | Treg | in vitro | LoF | 491 | Foxp3 |
| 33328215 | 2021 | Wang_CRISPRi_CART | human | CART | in vitro | LoF | 18904 | persistence |
| 30449619 | 2018 | Shifrut_CRISPRi_CD8T | human | CD8T | in vitro | LoF | 19114 | proliferation |
| 35750052 | 2022 | Belk_CRISPRi_CD8T | mouse | CD8T | in vitro | LoF | 18421 | persistence |
| 34879274 | 2021 | Zhao_CRISPRi_CD8T_invitro | mouse | CD8T | in vitro | LoF | 19642 | proliferation |
| 34879274 | 2021 | Zhao_CRISPRi_CD8T_invivo | mouse | CD8T | in vivo | LoF | 19642 | proliferation |
| 33328215 | 2019 | Ye_CRISPRi_CD8T_GBM | mouse | CD8T | in vivo | LoF | 1657 | infiltration |
| 38490212 | 2024 | Lin_CRISPRi_CD8T_persist | mouse | CD8T | in vitro | LoF | 19674 | persistence |
| 38490212 | 2024 | Lin_CRISPRi_CD8T_proliferate | mouse | CD8T | in vitro | LoF | 19674 | proliferation |
| 38490212 | 2024 | Lin_CRISPRi_CD8T_survival | mouse | CD8T | in vitro | LoF | 19674 | survival |
| 36002574 | 2022 | Carnevale_CRISPRi_Tcell | human | Tcell | in vitro | LoF | 19112 | proliferation |
| 36002574 | 2022 | Carnevale_CRISPRi_Treg | human | Treg | in vitro | LoF | 19112 | proliferation |
| 31827283 | 2019 | Wei_CRISPRko_CD8T | mouse | CD8+ T | in vivo | LoF | 3017 | infiltration |
| 35817986 | 2022 | Freimer_CRISPRko_CD4T_IL2RA | human | CD4+ T | in vitro | LoF | 1381 | IL2R |
| 35817986 | 2022 | Freimer_CRISPRko_CD4T_IL2 | human | CD4+ T | in vitro | LoF | 1381 | IL2 |
| 35817986 | 2022 | Freimer_CRISPRko_CD4T_CTLA4 | human | CD4+ T | in vitro | LoF | 1381 | CTLA4 |
| 34795452 | 2021 | Long_CRISPRko_Treg_metabolic | mouse | Treg | in vitro | LoF | 19666 | metabolic |
| 39663454 | 2025 | Arce_CRISPRi_CD4T_RestEff_IL2RA | human | CD4+ T | in vitro | LoF | 1349 | IL2RA |
| 39663454 | 2025 | Arce_CRISPRi_CD4T_StimEff_IL2RA | human | CD4+ T | in vitro | LoF | 1349 | IL2RA |
| 39663454 | 2025 | Arce_CRISPRi_Treg_IL2RA | human | Treg | in vitro | LoF | 1349 | IL2RA |
Bulk RNA-Seq Profiling T Cells Datasets
Chimeric antigen receptor (CAR) T Cells Datasets
| PMID | Accession | Year | CAR-T | #Respond | #No-Respond | Profiling method | ShinyApp |
|---|---|---|---|---|---|---|---|
| 33020644 | GSE151511 | 2020 | CD19 | 10 | 13 | scRNA-seq | Link |
| 34861191 | GSE160160 | 2021 | MSLN | 3 | 3 | scRNA-seq | Link |
| 37875502 | GSE243325 | 2023 | CD19/CD20 | 3 | 3 | scRNA-seq | Link |
| 37934007 | GSE246960 | 2024 | CD19 | 2 | 2 | scRNA-seq | Link |
| 37875502 | GSE223655 | 2023 | CD19/CD20 | 12 | 11 | bulkRNA-seq | N/A |
| 34861191 | GSE160154 | 2021 | MSLN | 9 | 13 | bulkRNA-seq | N/A |
| 35440691 | GSE147991 | 2022 | CD19 | 3 | 6 | bulkRNA-seq | N/A |
| 34861037 | GSE173918 | 2022 | CD19 | 3 | 3 | bulkRNA-seq | N/A |
| 34396987 | GSE175981 | 2021 | CD19 | 4 | 4 | bulkRNA-seq | N/A |
| 39151930 | GSE241456 | 2024 | CD19 | 3 | 3 | bulkRNA-seq | N/A |
| 37934007 | GSE245184 | 2024 | CD19 | 5 | 5 | bulkRNA-seq | N/A |
| 38408246 | GSE248382 | 2024 | CD19 | 3 | 3 | bulkRNA-seq | N/A |
T-cell Lymphoma/Leukemia Datasets
Autoimmune Diseases Transcriptomic Profiling Datasets
| DiseaseName | PMID | Accession | Year | #Disease | #Control | Profiling method |
|---|---|---|---|---|---|---|
| Systemic lupus erythematosus | 25861459 | GSE50772 | 2015 | 61 | 20 | Microarray |
| Systemic lupus erythematosus | 25736140 | GSE61635 | 2015 | 79 | 30 | Microarray |
| Systemic lupus erythematosus | 26382853 | GSE72509 | 2015 | 99 | 18 | RNA-seq |
| Inflammatory bowel disease | 31618209 | GSE126124 | 2019 | 59 | 32 | Microarray |
| Inflammatory bowel disease | 16436634 | GSE3365 | 2006 | 85 | 42 | Microarray |
| Inflammatory bowel disease | 29950172 | GSE112057 | 2018 | 75 | 12 | RNA-seq |
| Multiple sclerosis | 23895517 | GSE43591 | 2013 | 10 | 10 | Microarray |
| Multiple sclerosis | 22021740 | GSE21942 | 2011 | 14 | 15 | Microarray |
| Multiple sclerosis | 20190274 | GSE17048 | 2010 | 99 | 45 | Microarray |
| Multiple sclerosis | 28349074 | E-MTAB-5151 | 2017 | 49 | 27 | Microarray |
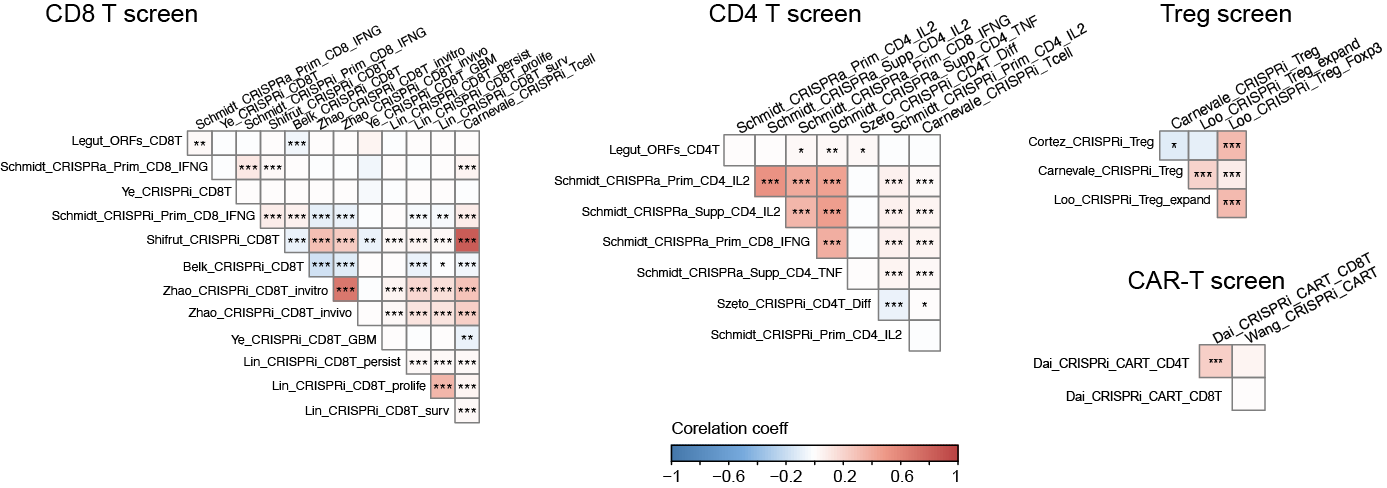
Correlations of T Cells CRISPR Screen Datasets
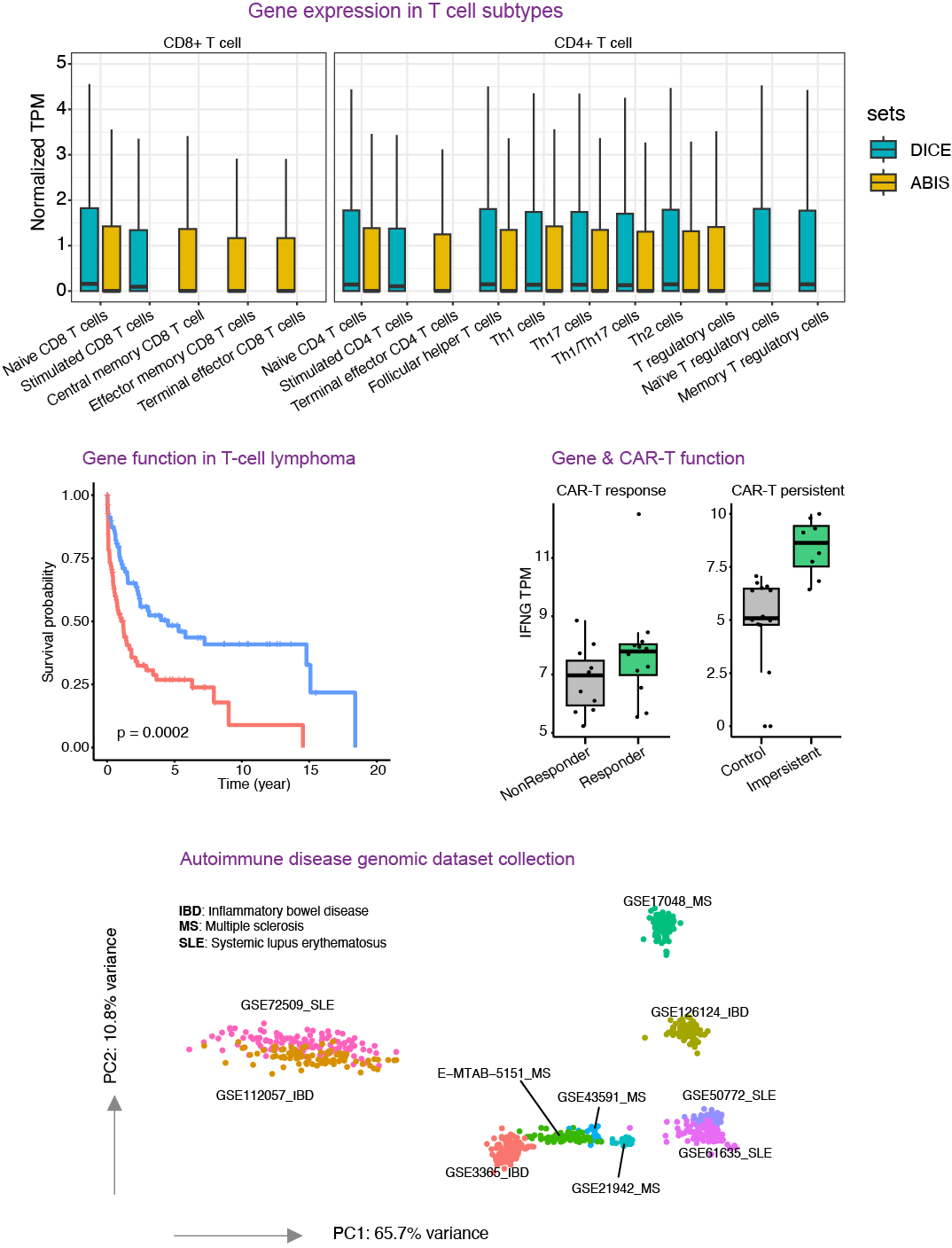
Summary of T cells Gene Transcriptomic Profiling Datasets
Copyright © Chen Lab, Yale School of Medicine, New Haven, CT 06516
Any comments and dataset suggestions, please contact us AT chuanpeng.dong@yale.edu